Image Segmentation using UNET Models
Note: Segmentation using Deep Learning requires the Deep Learning extension to the 2D Automated Analysis module. The Image-Pro Neural Engine must be installed. Installing the Image-Pro Neural Engine
Image-Pro supports image segmentation using UNET Models. UNET models perform semantic segmentation, assigning class labels to each pixel within a image. UNET is capable of assigning pixels into multiple classes. UNET was designed by Olaf Ronneberger, Philipp Fischer, and Thomas Brox of the University of Freiburg and was designed to work with very few training images and yield precise segmentation (Ronneberger, Fischer and Brox 2015). UNET can be used to segment arbitrarily large images by implementing an overlap-tile strategy.
https://lmb.informatik.uni-freiburg.de/people/ronneber/u-net/
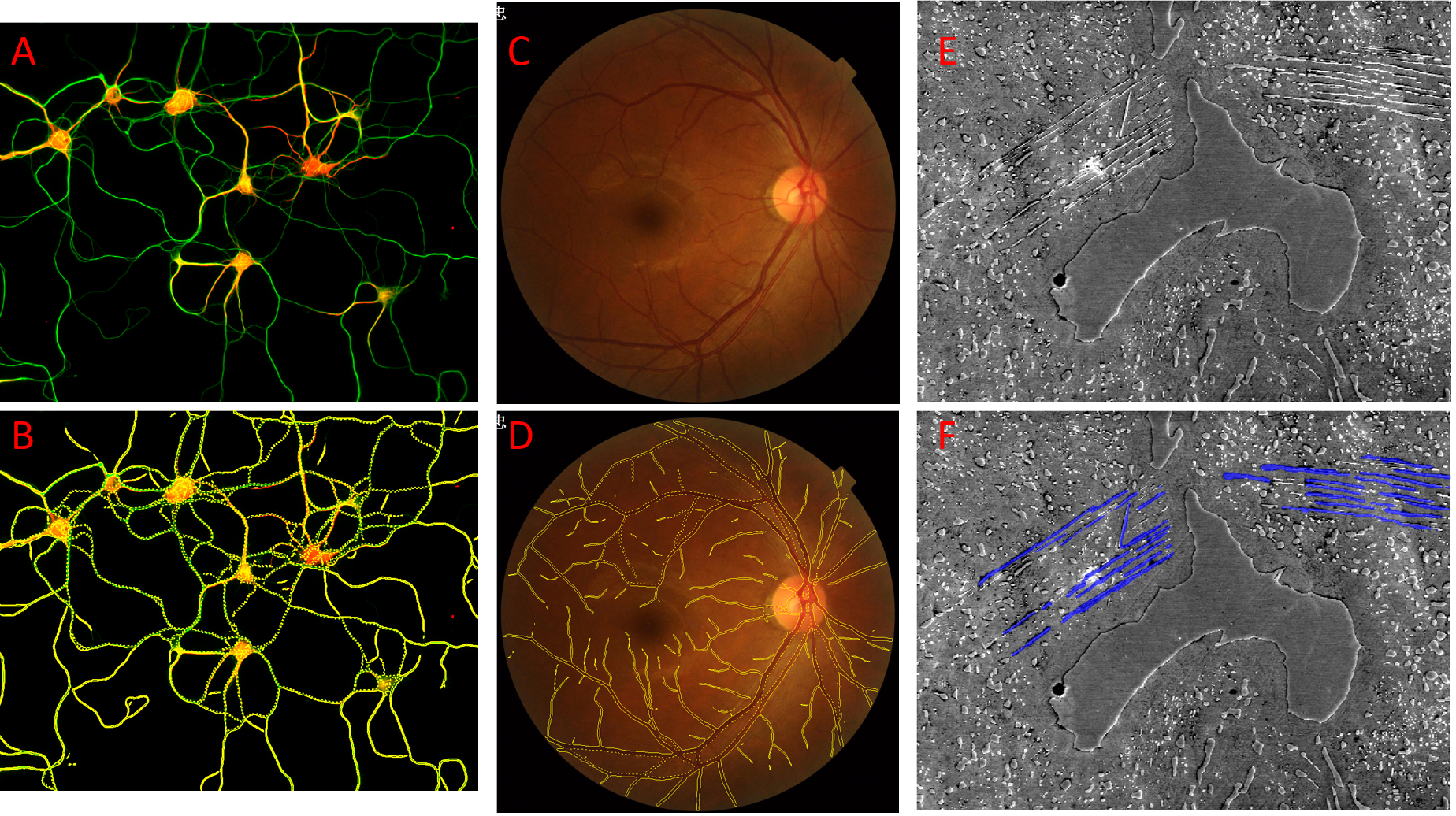
Figure showing semantic segmentation in Image-Pro using UNET models. Unlabeled images are shown in panels A, C, and E. The same images are shown with objects found by the Fluorescent Dendrites model in panel B, the Retinal Blood Vessels model in panel D, and the Windmanstatten Patterns model in Panel F.
To segment objects using UNET models in the active image select the AI button from the Segment tool group on the Count/Size ribbon. The AI Deep Learning Prediction panel will open.
Perform the following actions within the AI Deep Learning Prediction panel:
-
Load Model.
-
Click the Load button. The Open Model for Prediction dialog opens.
-
Select BaseUNET Semantic Segmentation from the Architecture drop down to show only UNET models.
-
Use the example images and descriptions to help you select a UNET model that's appropriate for your objects of interests and click Open.
-
-
Configure Classes
-
Name the classes and set the color and shape of predicted objects for each class.
-
-
Select Targets.
-
Select the channel that contains your objects of interest as the segmentation channel.
-
-
Set Model Options.
The are no options to set for UNET models, so skip this step and move on to Step 5.
-
Predict and Count.
-
Select the Show Output check box to automatically open the Deep Learning Output panel while prediction takes place. The Deep Learning Output panel will display information and feedback from the UNET model during prediction.
-
Select the Apply Filters/Splits check box if you wish to Count objects in the image with Ranges and automatic splits applied.
-
Click Count. Objects of interest that are found in your image will be displayed as measurement objects.
-
Use the Show/Hide button to toggle the display of objects on the image and to determine if you are happy that the measured objects are accurate.
-
Refining UNET Prediction
UNET models do not have settings that you can adjust.
If you fail to segment your images correctly with a UNET model, you can try:
-
Loading another more suitable model.
-
Refining your model with further training. Click Continue Training to open the model in the AI Deep Learning Training Panel.
-
Cloning the model and further training the cloned model. Click Clone Model to open the Clone Model dialog.
-
Make and train a new model.
Saving Deep Learning Segmentation Settings
It's convenient to save all of your deep learning settings including your model, model options, range settings, and split settings as a single settings file. To do this, click Save in the Measurements tool group on the Count/Size ribbon. A Save Measurements Options dialog opens. Name and save your measurements options (.iqo) file.
Restoring Saved Deep Learning Segmentation Settings
Click Load in the Measurements tool group on the Count/Size ribbon. A Load Measurements Options dialog opens. Navigate to your saved measurements options (.iqo) file and click Open. Your saved settings are restored.
Bibliography
Ronneberger, O., Fischer, P., Brox T.(2015) U-Net: Convolutional Networks for Biomedical Image Segmentation.
Medical Image Computing and Computer-Assisted Intervention (MICCAI), Springer, LNCS, Vol.9351: 234--241, 2015, available at arXiv:1505.04597 [cs.CV]
Learn more about all features in Count/Size